
TreeMatching is an R package that provides tools for matching trees between two spatial datasets, typically lidar-derived tree segmentations and field-measured datasets.
Matching trees between different surveys (e.g., lidar-derived and field-measured) is challenging due to non-rigid and local misalignments. This package introduces a robust approach where X and Y come from the spatial coordinates and Z is synthesized from tree DBH or tree height to improve the quality of the matching.
The current version includes one matching strategy based on solving Linear Sum Assignment Problem with the Hungarian algorithm.
Read the tutorial 📖
Example
library(TreeMatching)
data(PRF025_Field)
data(PRF025_Lidar)
PRF025_Field <- standardize(PRF025_Field, "Field_Xpj", "Field_Ypj", "DBH", "cm", crs = 2959)
PRF025_Lidar <- standardize(PRF025_Lidar, "X", "Y", "DBH", "m", crs = 2959)
center <- c(PRF025_Field$Easting[1], PRF025_Field$Northing[1])
treemap <- make_mapmatching(PRF025_Field, PRF025_Lidar, center, radius = 11.28)
treemap <- match_trees(treemap, method = lsap_matching, dxymax = 2, dzmax = 40)
plot(treemap, scale = 2)
plot(treemap, rgl = TRUE)
treemap$match_table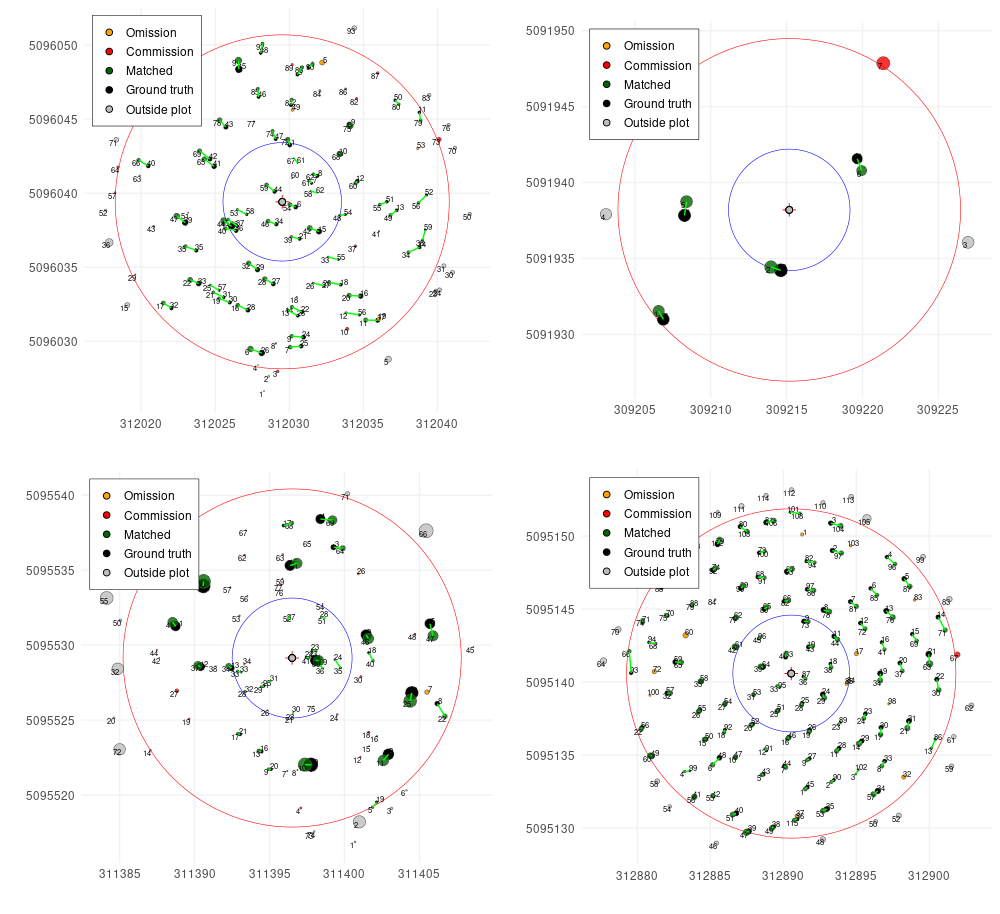
Sponsor
lidRalignment has been sponsored by the University of Sherbrooke

The development of the package was supported by data collected by Murray E. Woods and Margaret Penner (Forest Analysis Ltd) for a projected funded by the Forestry Futures Trust - KTTD program (Enhanced Forest Resource Inventory - forestryfutures.ca) with testing contribution by Bastien Vandendaele: Canadian Forest Service